HRD Harmonization Project
Homologous Recombination Deficiency Harmonization Project
Historically, cancer treatment involved and relied on chemotherapy, which attacks dividing cells throughout the body, including many cells that are not cancerous. In recent years, scientific innovation has allowed for the safe and effective use of targeted therapies, which can precisely attack cancer cells that have specific signals known as biomarkers, potentially providing more effective and safer treatments for patients. Since not all cancer cells have specific biomarkers targetable by current therapies, it is necessary to identify which patients have the biomarker and may benefit from specific targeted therapies. To help identify which patients have a specific biomarker, diagnostic developers have created tests, also known as assays.
Test developers have different approaches for how they identify a specific biomarker increasing variability across results from different tests. One such biomarker facing issues with harmonization is homologous recombination deficiency (HRD), which is used to determine which patients with certain types of cancer should be treated with a drug called a PARP inhibitor. A combination of factors is used to determine HR status, and different assays, or tests, use different approaches. This lack of harmonization can make it challenging for patients and providers to interpret the results of these tests.
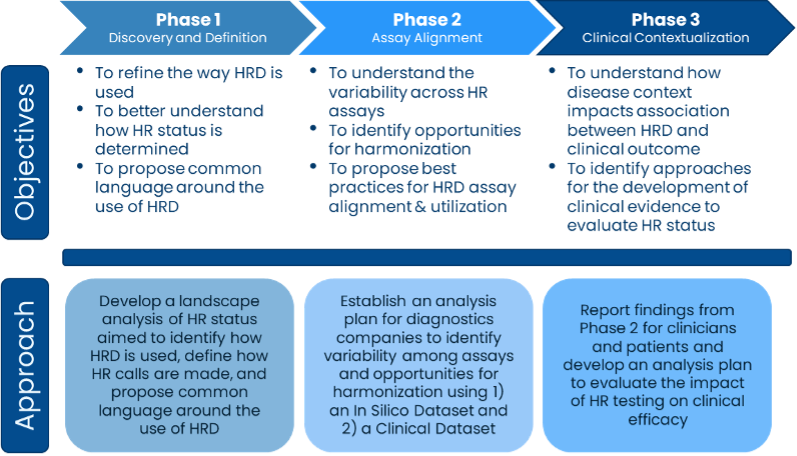
To understand the variability in assays that test for HRD and determine solutions to harmonize the assays, Friends convened a group of test developers, patient advocates, providers, and government officials to address concerns about the lack of consistency in identifying HRD, its prognostic value for how well a treatment might work, and its use as a predictive biomarker for how a patient’s prognosis might differ due to the presence of the biomarker. During the first phase of the project, the group conducted a landscape analysis of how HRD is used and defined to establish a baseline understanding of inconsistencies across tests and to propose common terminology.
Phase 2 of the project works to align assays that measure HRD. The group identified samples that each test developer involved in the project could analyze using their HRD assay and then report the HRD status, or whether the sample has HRD or not. The HR status will be compared across assays to understand the level of agreement among the assays. These results will be used to work towards defining where there are differences across assays and inform solutions to harmonize the results.
Friends’ HRD Harmonization Project will help determine the amount of agreement among assays that measure HRD and propose solutions to improve agreement in the future. If assay developers use agreed upon solutions in their assays, providers and patients will have more consistent results regardless of which assay is used to measure HRD. Without Friends coordination and support from collaborative sponsors, groups may never align on a solution to improve consistency across assays and interpreting results from assays would be more challenging for patients and providers.
Project Timeline
Project Outcomes
2024
- Friends hosted a public meeting sharing the full findings from the HRD Harmonization Project in February.
2023
- The Clinical Sample Findings that were presented at AACR Special Conference: Ovarian Cancer in October showed moderate variability in HRD status calls.
2022:
- A manuscript describing opportunities and best practices to align the definition of HRD and the parameters that contribute to the determination of HRD status was published in The Oncologist.
- Friends launched the HRD Harmonization Project.
- Initial findings from the In Silico Findings from the project were presented at the AMP 2022 Annual Meeting and Expo in November demonstrating the inter-assay agreement on HRD status calls was variable.
2021
- The HRD working group convened and characterized current and future uses of HRD and HRD assays in oncology drug development.
Project Partners
Phase 1: Agilent Technologies, Ambry Genetics, Arizona State University, AstraZeneca, Bristol Myers Squibb, Caris Life Sciences, EMD Serono, Inc. (Merck KGaA), Foundation Medicine, Inc., GlaxoSmithKline, Guardant Health, Inc., Janssen Pharmaceuticals, Merck & Co., Inc. (MSD), Myriad Genetics, the National Cancer Institute (NCI), Pfizer, Tempus Labs, Inc., Thermo Fisher Scientific, the U.S. Food and Drug Administration (FDA), University of Alabama at Birmingham, and the University of Heidelberg
Phase 2: ACT Genomics, AmoyDx, AstraZeneca, Bayer, Bionano Genomics, Inc., BostonGene Corporation, Bristol Myers Squibb, Burning Rock, Diagnostic Laboratory Services, Inc., DNAnexus, EMD Serono, Inc., European Organisation for Research and Treatment of Cancer (EORTC), Foundation Medicine, Inc., Genentech, Inc., Guardant Health, Inc., Illumina, Inc., Invitae Corp., Laboratory Corporation of America (Labcorp), MD Anderson Cancer Center, Merck & Co., Inc. (MSD), Molecular Characterization Laboratory (MoCha) at Frederick National Laboratory, the National Cancer Institute (NCI), Neogenomics Laboratories, Inc., PathAI, Pfizer, Pillar Biosciences, SOPHiA Genetics, Tempus AI Inc., Thermo Fisher Scientific, the U.S. Food and Drug Administration (FDA), University of Alabama at Birmingham, and the University of Heidelberg
